New Approach - Structural Features#
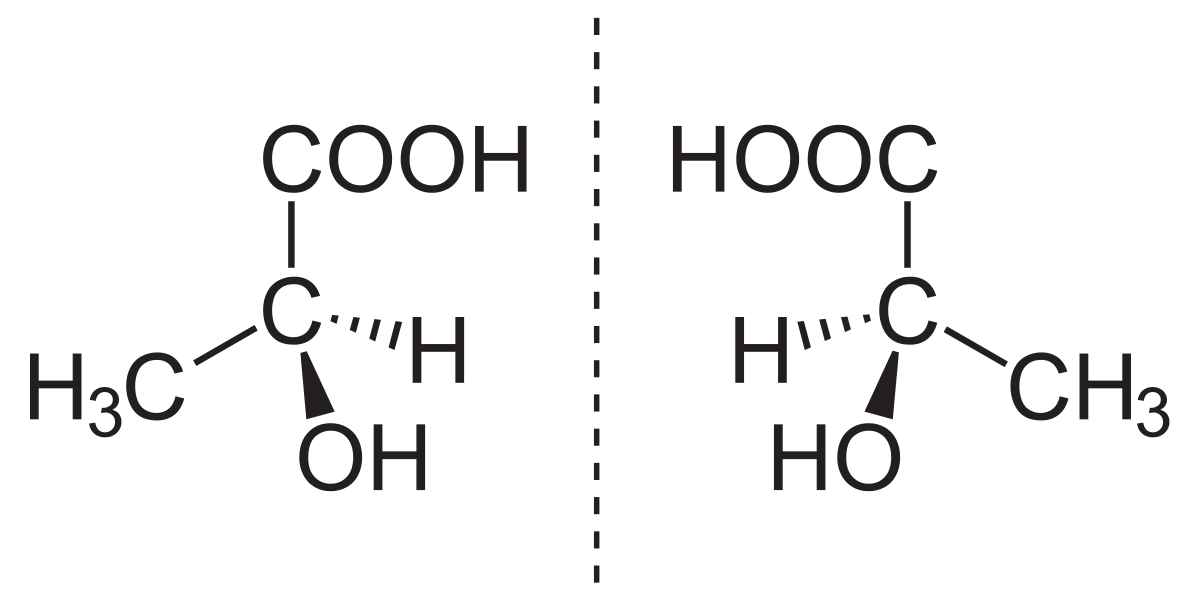
The old approach could not detect a difference within the pair because molecular descriptors were used as features.
So our apporach here was to use the physicochemical, or structural, characterisitcs as features to predict divergence. We know the strcutures of molecules in an enantioemric pair are different since they are rotated versions of each other.
We collected a dataset of 174 enantiomeric pairs and we used the Cononical SMILES[1] string of each molecule to compute the strcutural features for that molecule.
The Python packages, Mordred and Morgan, were used to compute strcutural features from the SMILES strings.